To generate acceptable files, please use alignment program BLAT with option -out=pslx.
Please edit the database information by click on the pencil icon next to your dataset. Select the corresponding genome build.
What it does
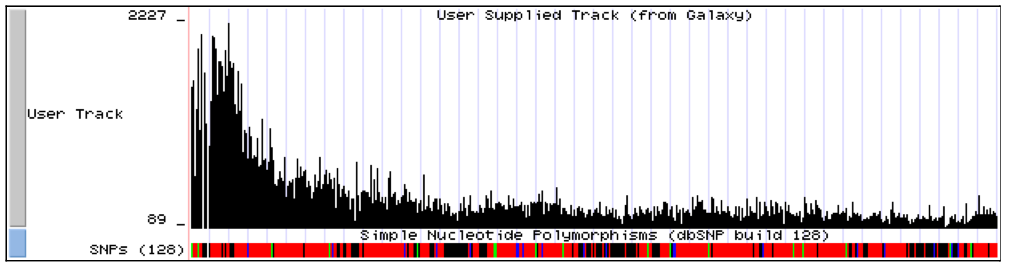
This tool takes BLAT pslx output and returns a wig-like file showing the number of reads (coverage) mapped at each chromosome location. Use Graph/Display Data --> Build custom track tool to show the coverage mapping in UCSC Genome Browser.
Example
Showing reads coverage on human chromosome 22 (partial result) in UCSC Genome Browser Custom Track: