Mercurial > repos > davidecangelosi > pipe_t
comparison README.md @ 30:30ab487171ab draft
planemo upload for repository https://github.com/igg-molecular-biology-lab/pipe-t.git commit 488b3c091e8bf80f741a3e46b929427f1658ba3a-dirty
| author | davidecangelosi |
|---|---|
| date | Thu, 10 Oct 2019 04:24:43 -0400 |
| parents | 46cb147d53df |
| children |
comparison
equal
deleted
inserted
replaced
| 29:8cb95051067d | 30:30ab487171ab |
|---|---|
| 90 | 90 |
| 91 The Galaxy Admin User has the username `admin@galaxy.org` and the password `1234`. In order to use some features of Galaxy, like import history, one has to be logged in with this username and password. | 91 The Galaxy Admin User has the username `admin@galaxy.org` and the password `1234`. In order to use some features of Galaxy, like import history, one has to be logged in with this username and password. |
| 92 | 92 |
| 93 Docker images are "read-only", all your changes inside one session will be lost after restart. This mode is useful to present Galaxy to your colleagues or to run workshops with it. To install Tool Shed repositories or to save your data you need to export the calculated data to the host computer. | 93 Docker images are "read-only", all your changes inside one session will be lost after restart. This mode is useful to present Galaxy to your colleagues or to run workshops with it. To install Tool Shed repositories or to save your data you need to export the calculated data to the host computer. |
| 94 | 94 |
| 95 Run this command setting your local landing path (`/host/path/targetfolder/`): | 95 Run this command: |
| 96 ``` | 96 ``` |
| 97 docker run -d -p 8080:80 \ | 97 docker create -v /export \ |
| 98 --name pipe-t-store \ | |
| 99 davidecangelosi/galaxy-pipe-t \ | |
| 100 /bin/true | |
| 101 ``` | |
| 102 and then run: | |
| 103 ``` | |
| 98 docker run -d -p 21:21/tcp -p 443:443/tcp -p 80:80/tcp -p 8800:8800/tcp -p 9002:9002/tcp \ | 104 docker run -d -p 21:21/tcp -p 443:443/tcp -p 80:80/tcp -p 8800:8800/tcp -p 9002:9002/tcp \ |
| 99 -v /host/path/targetfolder/:/export/ \ | 105 --volumes-from pipe-t-store \ |
| 100 davidecangelosi/galaxy-pipe-t:latest | 106 davidecangelosi/galaxy-pipe-t:latest |
| 101 ``` | 107 ``` |
| 102 | 108 |
| 103 | 109 |
| 104 For more information about the parameters and docker usage, please refer to https://github.com/bgruening/docker-galaxy-stable/blob/master/README.md#Usage | 110 For more information about the parameters and docker usage, please refer to https://github.com/bgruening/docker-galaxy-stable/blob/master/README.md#Usage |
| 159 - A tab-separated text file containing data after imputation | 165 - A tab-separated text file containing data after imputation |
| 160 - A tab-separated text file containing the results of the differential expression analysis. | 166 - A tab-separated text file containing the results of the differential expression analysis. |
| 161 | 167 |
| 162 ### Example application <a name="first-example" /> [[toc]](#toc) | 168 ### Example application <a name="first-example" /> [[toc]](#toc) |
| 163 | 169 |
| 164 In this example, we will show you how to perform a simple analysis with PIPE-T on mCRC data. We have chosen the 53 metastatic colorectal cancer study that we presented in our manuscript. | 170 In this example, we will show you how to perform a simple analysis with PIPE-T on mCRC data. We have chosen the 53 metastatic colorectal cancer study whose gene expression profile was available in the Gene Expression Omnibus with accession GSE52513. |
| 165 | 171 |
| 166 | 172 |
| 167 The top rows relative to an example file of a hypothetical ListOfFile is reported in the table below. Note that file contains data about the type of the file, dates and any other relevant information about the experiment. | 173 The top rows relative to an example file of a hypothetical ListOfFile is reported in the table below. Note that file contains data about the type of the file, dates and any other relevant information about the experiment. |
| 168 | 174 |
| 169 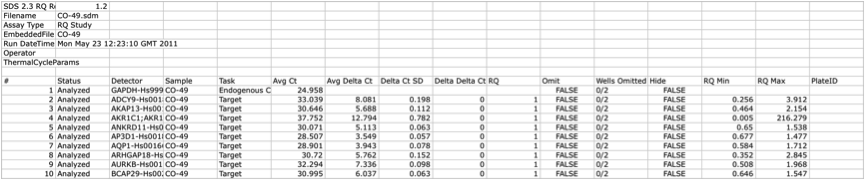 | 175  |
