Author(s) Ron Wehrens (ron.wehrens@gmail.com), Georg Weingart, Fulvio Mattivi
Galaxy wrapper and scripts developpers Guitton Yann LABERCA yann.guitton@oniris-nantes.fr and Saint-Vanne Julien julien.saint-vanne@sb-roscoff.fr
Please cites
metaMS : Wehrens, R.; Weingart, G.; Mattivi, F. Journal of Chromatography B.
xcms : Smith, C. A.; Want, E. J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. Anal. Chem. 2006, 78, 779–787.
CAMERA : Kuhl, C.; Tautenhahn, R.; Böttcher, C.; Larson, T. R.; Neumann, S. Analytical Chemistry 2012, 84, 283–289.
metaMS.runGC
Description
metaMS.runGC is a function dedicated to GCMS data processing from converted files to the generation of pseudospectra (compounds) table. Current version for metaMS R package: 1.18.1
Process: Each of the converted data (cdf, mzML...) is profiled by a combination of xcms and CAMERA functions. Then all the mass spectra of detected peaks are compared and clustered. For more details see metaMS : Wehrens, R.; Weingart, G.; Mattivi, F. Journal of Chromatography B (10.1016/j.jchromb.2014.02.051) link
Main outputs: A PeakTable is generated with one line per "compound" and one column per sample. A dataMatrix.csv file is generated and can be used for PCA or for further analysis. A peakspectra.mps file is generated that contains all the spectra of the detected compounds in MSP format. That file can be used for database search online (Golm, MassBank) or locally (NIST MSSEARCH) for NIST search a tutorial is available here.
Workflow position
Upstream tools
You can start from here (XCMS 1.x) or use result file from (XCMS 3.x) :
| Name | output file | format | parameter |
|---|---|---|---|
| xcms.xcmsSet | xset.RData RData | RData | file |
Downstream tools
| Name | Output file | Format |
|---|---|---|
| Determine Vdk or Lowess | dataMatrix.tsv | Tabular |
| Normalization Vdk/Lowess | dataMatrix.tsv | Tabular |
| Anova | dataMatrix.tsv | Tabular |
| PCA | dataMatrix.tsv | Tabular |
| Hierarchical Clustering | dataMatrix.tsv | Tabular |
| Golm Metabolome Search | peakspectra.msp | Text |
General schema of the metabolomic workflow for GCMS
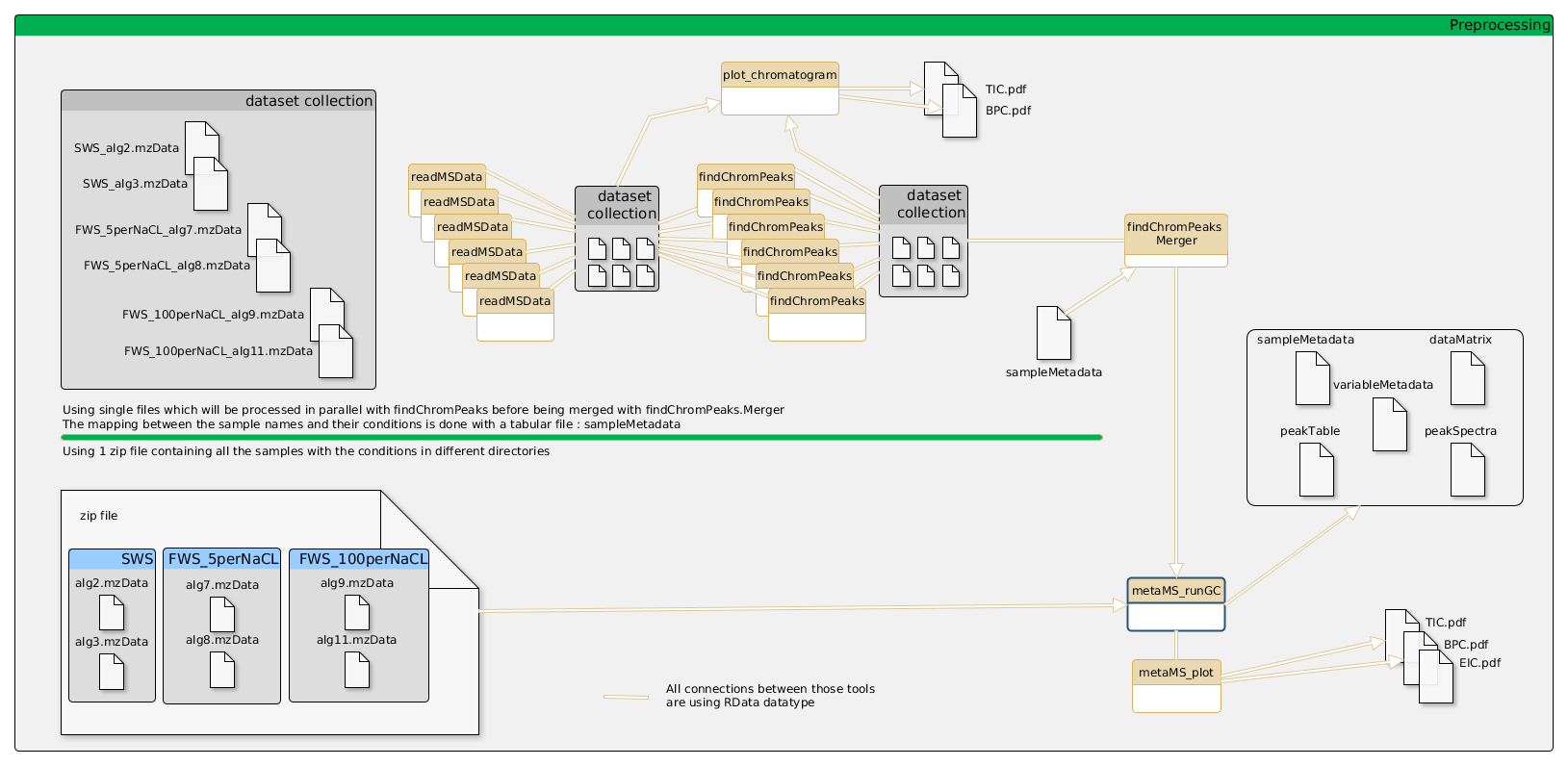
Input files
If you choose to use results from xcms.xcmsSet with XCMS > 3.0
| Parameter : num + label | Format |
|---|---|
| 1 : RData file from XCMS | RData |
Or converted GCMS files (mzML, CDF...) in your local libray with XCMS < 3.0 (from metaMS)
Parameters
Parameters are described in metaMS R package and mainly correspond to those of xcms.xcmsSet
Outputs
- The output file dataMatrix.tsv is a tabular file. You can continue your analysis using it in the statistical tools.
- The output file peakspectra.msp is a text file. You can continue your analysis using it in the golm search tool. Or you can load it in your personnal NIST MSsearch program (c:/NISTXX/mssearch/nistms.exe) that tool is in general installed by default on GCMS apparatus. Tutorial available here.
Working Example
Reference Data for testing are taken from:
Dittami,S.M. et al. (2012) Towards deciphering dynamic changes and evolutionary mechanisms involved in the adaptation to low salinities in Ectocarpus (brown algae): Adaptation to low salinities in Ectocarpus. The Plant Journal
Input files
RData file from XCMS -> xset_merged.RData
Parameters
Settings -> DefaultDB option -> show | DB file: -> W4M0004_database_small.msp...all default option values
Output files
RData file -> rungc.RData
Changelog/News
Version 3.0 - 20/05/2019
- Divided tool into two new. One will be metaMS_plot and this one stay the same without plotting chromatograms and without zip files
Version 2.1 - 08/06/2017
- Quality: add sessionInfo logs with packages versions
- Processing: add RI usage
Version 1.1 - 11/07/2016
- TEST: refactoring to pass planemo test using conda dependencies
Version 1.0 - 01/06/2015
- NEW: new tool