Contact matrix plot
hicPlotMatrix is a visualization tool for Hi-C contact matrices. It supports to plot genome-wide contact matrices, one or multiple chromosomes, a region or two regions against each other. Additionally it can plot the result of a principal component analysis obtained by hicPCA to have a better understanding of A / B compartments.
Usage
This tool can be used on any h5 or cool Hi-C contact matrix. It is noteworthy that for comparisons of 2 matrices or more, they must all have the same or similar number of contacts.
Output
hicPlotMatrix outputs a heatmap of a contact matrix in either png or svg format, below is an example of such a plot:
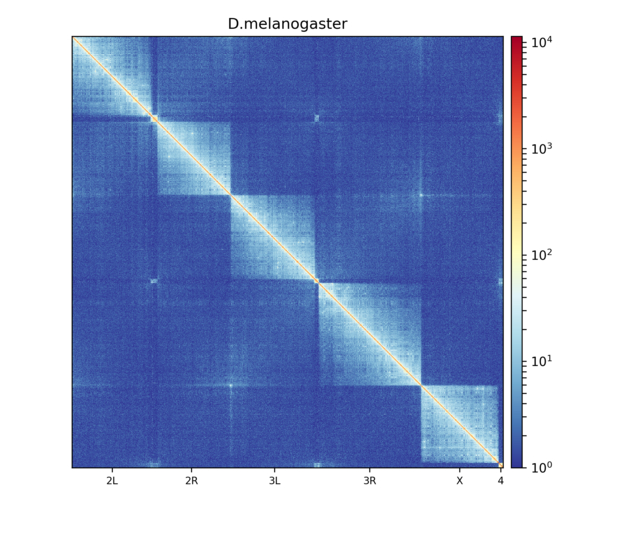
Contact matrix of Drosophila melanogaster embryos visualized using hicPlotMatrix. Hi-C matrix bins were merged to a 25 kb bin size using hicMergeMatrixBins and the matrix has then been corrected using hicCorrectMatrix before plotting.
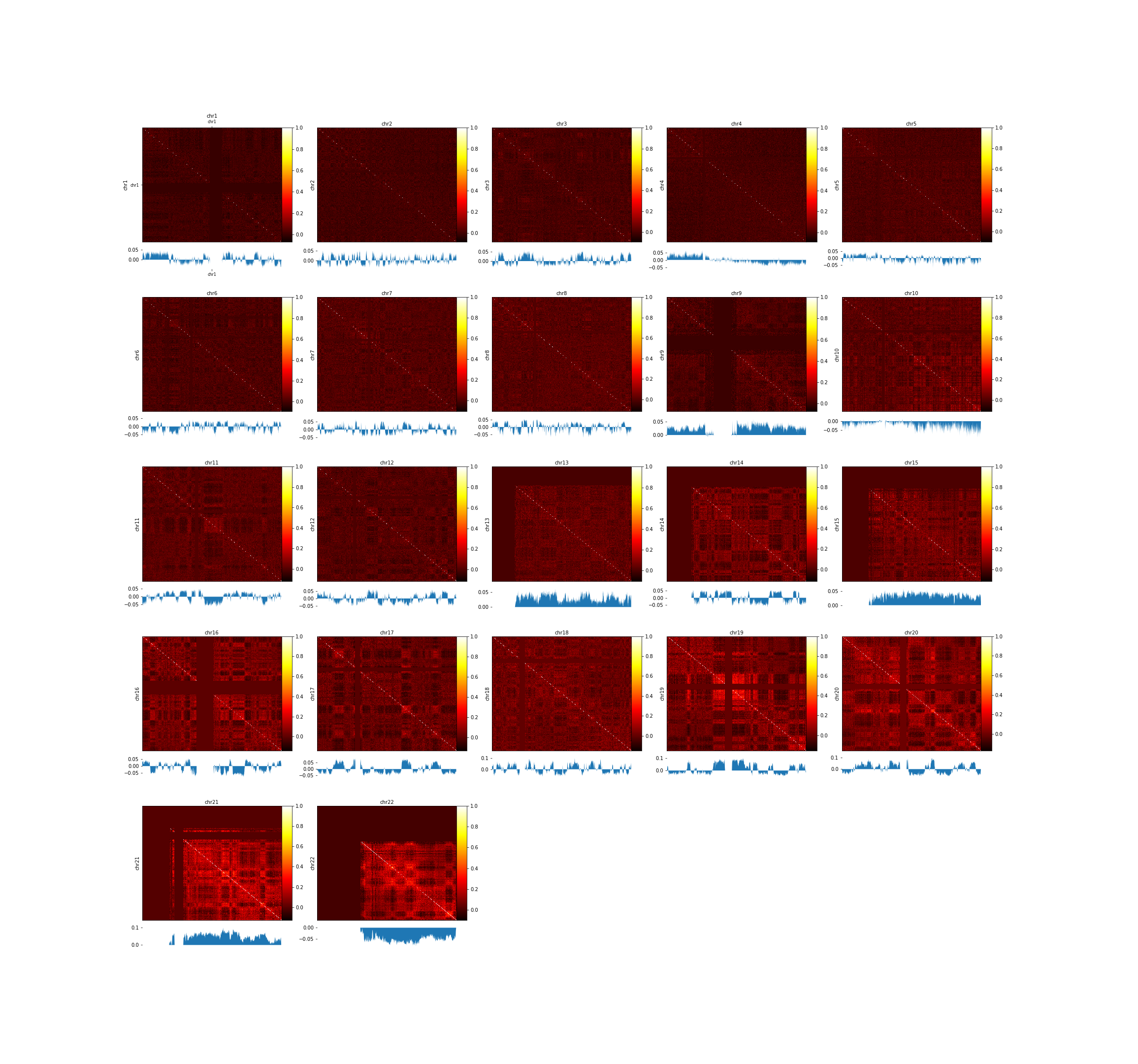
Another example is available below using Hi-C data published by Lieberman-Aiden in 2009, (GSE18199). A Hi-C contact matrix has been plotted with the --perChr option and the first eigenvector (pca1) computed by hicPCA. For this plot a pearson correlated matrix was used, which is computed by first creating an observed / expected matrix and then a pearson correlation matrix with hicTransform.