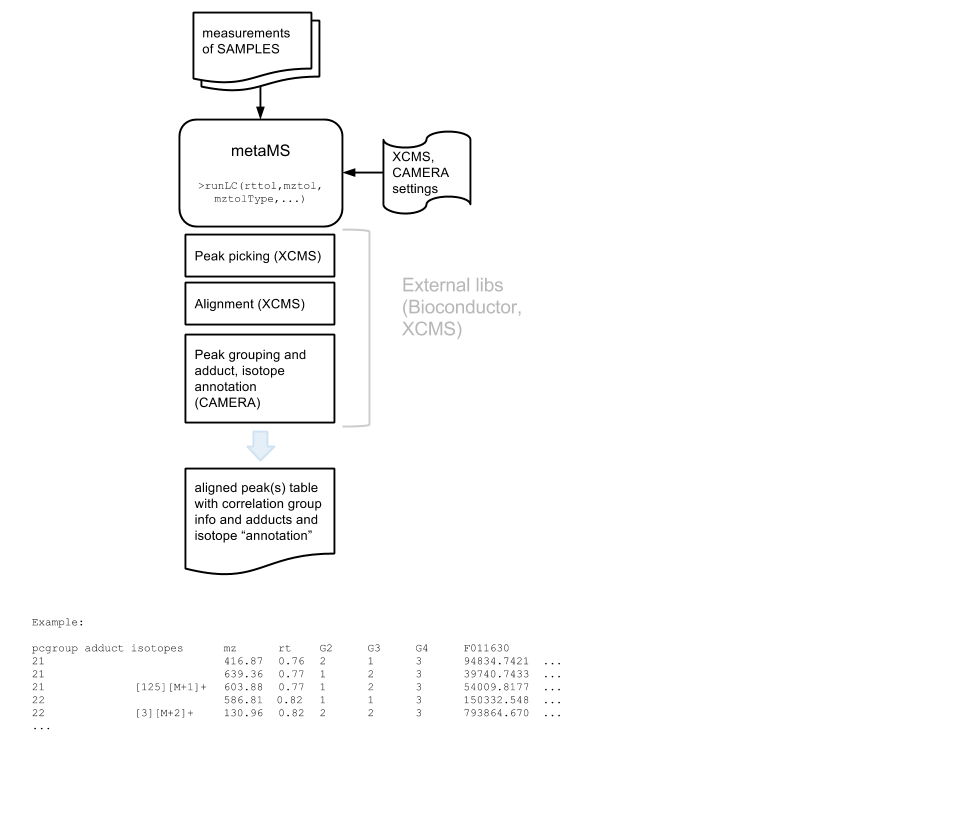
Runs metaMS process for LC/MS feature feature picking, aligning and grouping. This part of the metaMS process makes use of the XCMS and CAMERA tools and algorithms. CAMERA is used for automatic deconvolution/annotation of LC/ESI-MS data. The figure below shows the main parts of the metaMS process wrapped by this tool.
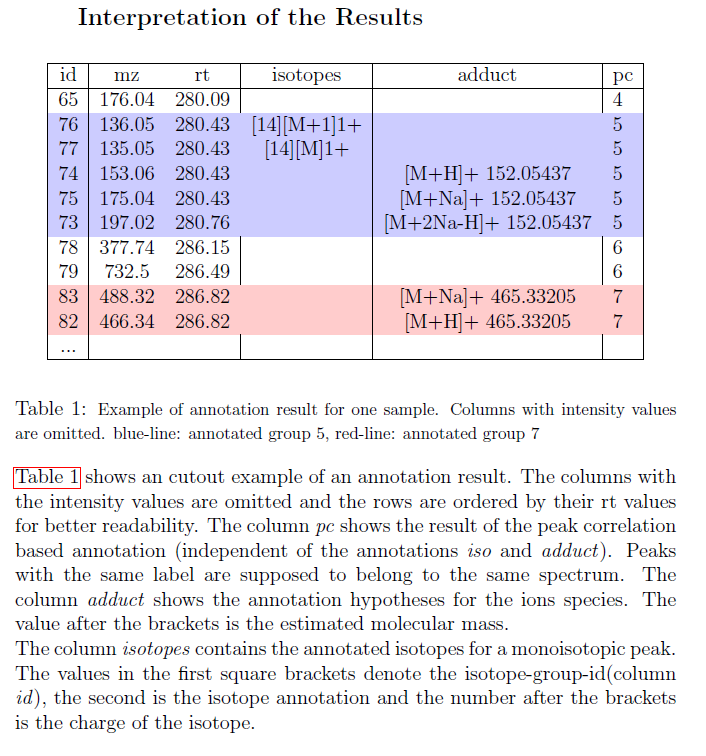
From CAMERA documentation:
References
If you use this Galaxy tool in work leading to a scientific publication please cite the following papers:
Wehrens, R.; Weingart, G.; Mattivi, F. (2014). metaMS: an open-source pipeline for GC-MS-based untargeted metabolomics. Journal of chromatography B: biomedical sciences and applications, 996 (1): 109-116. doi: 10.1016/j.jchromb.2014.02.051 handle: http://hdl.handle.net/10449/24012
Wrapper by Pieter Lukasse.