Authors Eric Fontanillas created the version 1 of this pipeline. Victor Mataigne developped version 2.
Galaxy integration Julie Baffard and ABiMS TEAM, Roscoff Marine Station
Contact support.abims@sb-roscoff.fr for any questions or concerns about the Galaxy implementation of this tool.Credits : Gildas le Corguillé, Misharl Monsoor
Description
This tool takes files containing nucleic aligned sequences and search the ORF and the CDS.
Inputs
Input files : (multiple) fasta files with nucleic aligned sequences.
Parameters
- methionine : choose to consider the methionine in the search of CDS.
- yes/no.
- 'Minimal number of species in each locus'
- Default : 10 (integer).
- 'min_length_seq' :
- minimal length of the sequence (in amino acids). when the removal of the indel is done, the minimal length equals : previous length - 20. for example if you choose 50 for the minimal length, the actual length equals 30. Default : 50 (integer).
- 'min_length_subseq' :
- minimal length of the subsequence (in amino acids). subsequence means the part of the original sequence between 2 sets of indels. an indel set is composed by more than 2 indels, if not the set is considered as unknown amino acid. Default : 15 (integer).
- 'min_length_nuc' :
- Minimal length of the sequence in the nucleic format, without indels. Default : 50 (integer).
- others parameters allowing to choose which outputs you desire :
- outputs with best ORFs.
- outputs with CDS, with or without indels.
- in proteic or nucleic format.
Outputs
- ORF_Search
- the log file (mainly statistics about the tool).
- ORF_Search_Best_ORF_aa
- the output with the best ORF in the proteic format.
- ORF_Search_Best_ORF_nuc
- the output with the best ORF in the nucleic format.
- ORF_Search_CDS_aa
- the output with the CDS (regardless the Methionine) in the proteic format.
- ORF_Search_CDS_nuc
- the output with the CDS (regardless the Methionine) in the nucleic format.
- ORF_Search_CDS_with_M_aa
- the output with the CDS (considering the Methionine) in proteic format. the rule : they must have a methionine before the minimal length of the sequence. for example before the 30 last amino acid.
- ORF_Search_CDS_with_M_nuc
- the output with the CDS (considering the Methionine) in nucleic format. the rule : they must have a methionine before the minimale length of the sequence. for example before the 30 last amino acid.
- ORF_Search_CDS_without_indel_aa
- is the output with the CDS without indel in proteic format. considering the Methionine or not : according to the option chosen.
- ORF_Search_CDS_without_indel_nuc
- is the output with the CDS without indel in proteic format. considering the Methionine or not : according to the option chosen.
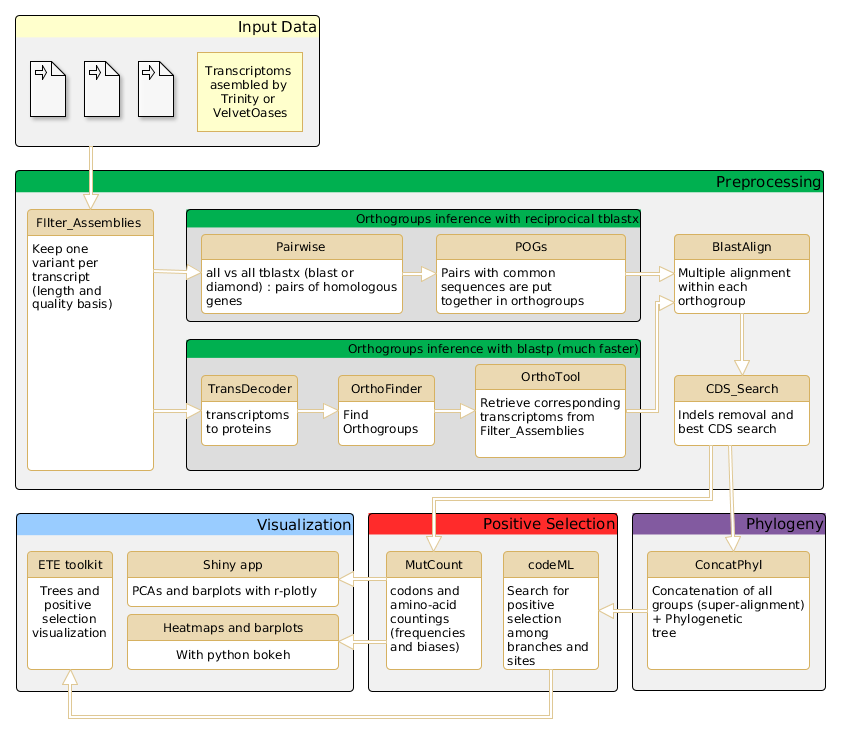
The AdaptSearch Pipeline
Changelog
Version 2.0 - 05/07/2017
- NEW: Replace the zip between tools by Dataset Collection
Version 1.0 - 13/04/2017
- Added functional test with planemo
- planemo test with conda dependency for python
- Scripts renamed + symlinks to the directory 'scripts'