Description
Get the counts of sequences per sample for the different stages of the dada pipeline.
Usage
Inputs:
Any number of results of dada2 steps in the following form:
- a collection of results from dada, mergePairs, or the collection of statistics from filterAndTrim (the identifiers of the collection elements are used as sample names)
- the results of dada in non-batch mode
- the result of makeSequenceTable or removeBimeraDenovo
Output:
A table containing the number of sequences per sample (rows) for each input (columns)
Details
For results from
- dada, and mergePairs the sum of the result of dada2's getUniques function is used
- makeSequenceTable, and removeBimeraDenovo R's rowSums function is used
Overview
The intended use of the dada2 tools for paired sequencing data is shown in the following image.
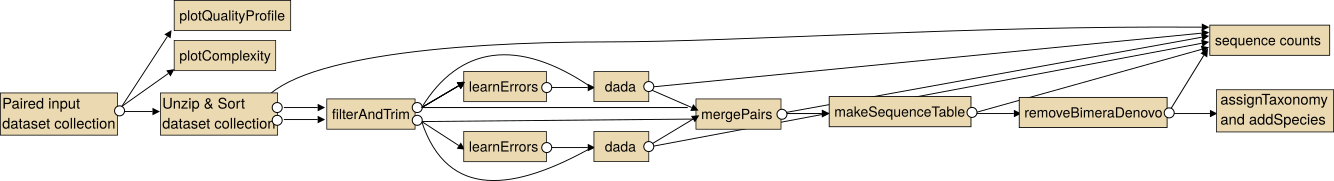
Note: In particular for the analysis of paired collections the collections should be sorted lexicographical before the analysis.
For single end data you the steps "Unzip collection" and "mergePairs" are not necessary.
More information may be found on the dada2 homepage:: https://benjjneb.github.io/dada2/index.html (in particular tutorials) or the documentation of dada2's R package https://bioconductor.org/packages/release/bioc/html/dada2.html (in particular the pdf which contains the full documentation of all parameters)