
What it does
Add taxonomic affiliation in abundance file.
Inputs/outputs
Inputs
Sequence file:
The sequences (format FASTA).
Abundance file:
The abundance of each OTU in each sample (format BIOM).
Outputs
Abundance file (tax_affiliation.biom):
The abundance file with affiliation (format BIOM).
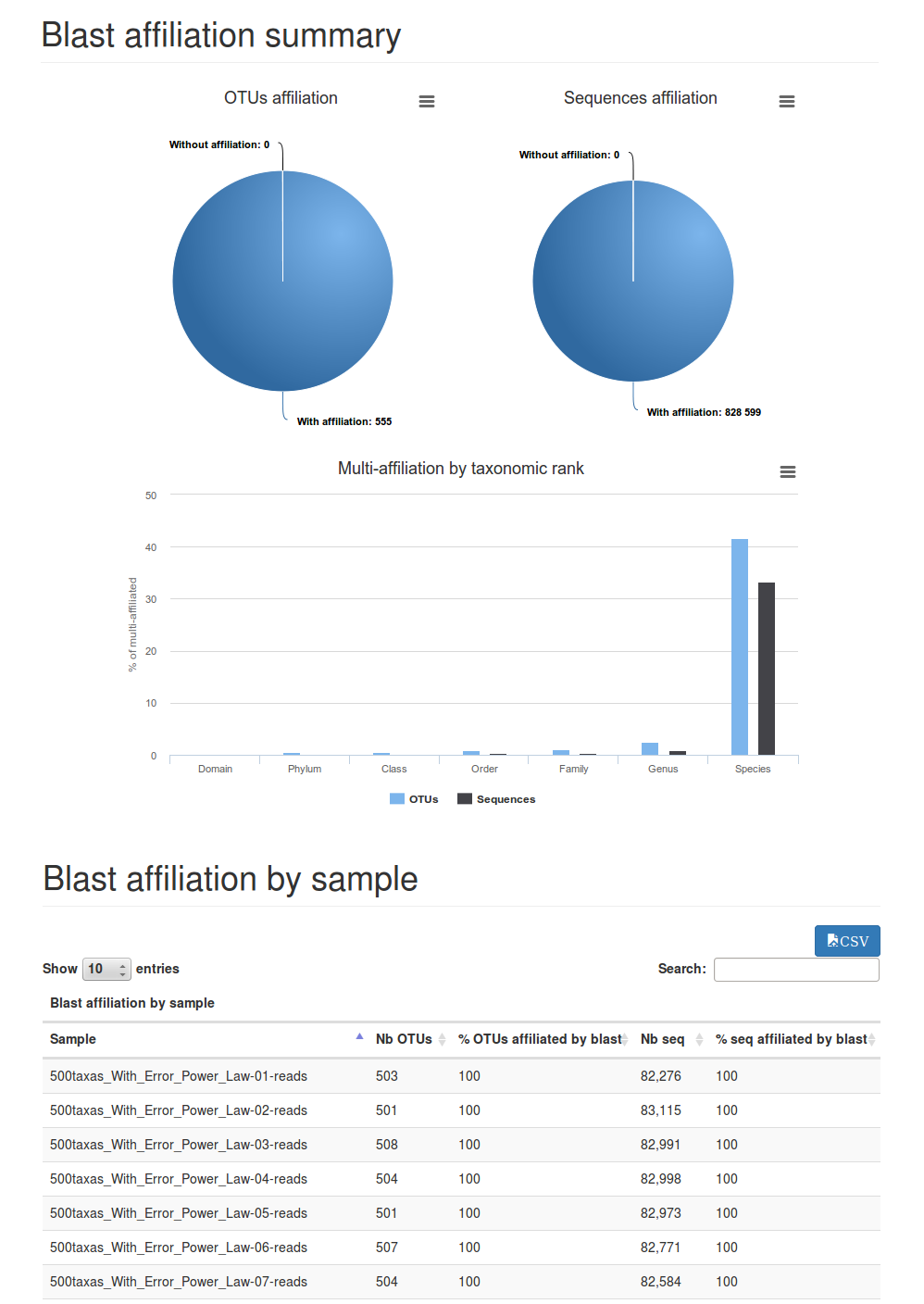
Summary file (report.html):
This file presents the number of sequences affiliated by blast, and the number of multi-affiliation (format HTML).
Reference database
The databases come from SILVA a comprehensive and quality checked rRNA database.
FROGS database(s) pre-process:
- The non-redondant small sub-unit database is retrieved from SILVA.
- This database is processed to keep only the taxonomy level: Domain - Phylum - Class - Order - Family - Genus - Species
- The database is splitted in two databses: prokaryote and eukaryote.
How it works
| Steps | Description |
|---|---|
| 1 | RDPClassifier is used with database to associate to each OTU a taxonomy and a bootstrap (example: Bacteria;(1.0);Firmicutes;(1.0);Clostridia;(1.0);Clostridiales;(1.0);Clostridiaceae 1;(1.0);Clostridium sensu stricto;(1.0);). |
| 2 | blastn+ is used to find alignment between each OTU and the database. Only the bests hits with the same score has reported. |
| 3 | For each OTU with several blastn+ results a consensus is determined on each taxonomic level. If all the taxa in a taxonomic rank are identical the taxon name is reported otherwise Multi-affiliation is reported. By example, if you have an OTU with two corresponding sequences, the first is a Bacteria;Proteobacteria;Gamma Proteobacteria;Enterobacteriales, the second is a Bacteria;Proteobacteria;Beta Proteobacteria;Methylophilales, the consensus will be Bacteria;Proteobacteria;Multi-affiliation;Multi-affiliation. |
Advices
This tool can take large time. It is recommended to filter your abundance and your sequence file before this tool (see FROGS Filters).
As you can see the affiliation of each OTU is not human readable in outputed abundance file. We provide a tools to convert these BIOM file in tabulated file, see the FROGS BIOM to TSV tool.
Contact
Contacts: frogs@inra.fr
Repository: https://github.com/geraldinepascal/FROGS
Please cite the FROGS Publication: Escudie F., Auer L., Bernard M., Cauquil L., Vidal K., Maman S., Mariadassou M., Hernadez-Raquet G., Pascal G., 2015. FROGS: Find Rapidly OTU with Galaxy Solution. In: The environmental genomic Conference, Montpellier, France, http://bioinfo.genotoul.fr/fileadmin/user_upload/FROGS_2015_GE_Montpellier_poster.pdf
Depending on the help provided you can cite us in acknowledgements, references or both.