This tool exports an AnnData dataset to a Loom file (write_loom method) or a Tabular file (write_csvs method)
It can also create a series of tabular files from an input loom dataset.
AnnData
AnnData provides a scalable way of keeping track of data together with learned annotations. It is used within Scanpy, for which it was initially developed.
AnnData stores a data matrix X together with annotations of observations obs, variables var and unstructured annotations uns.
AnnData stores observations (samples) of variables (features) in the rows of a matrix. This is the convention of the modern classics of statistics (Hastie et al., 2009) and machine learning (Murphy, 2012), the convention of dataframes both in R and Python and the established statistics and machine learning packages in Python (statsmodels, scikit-learn).
More details on the AnnData documentation
Loom data
Loom files are an efficient file format for very large omics datasets, consisting of a main matrix, optional additional layers, a variable number of row and column annotations, and sparse graph objects.
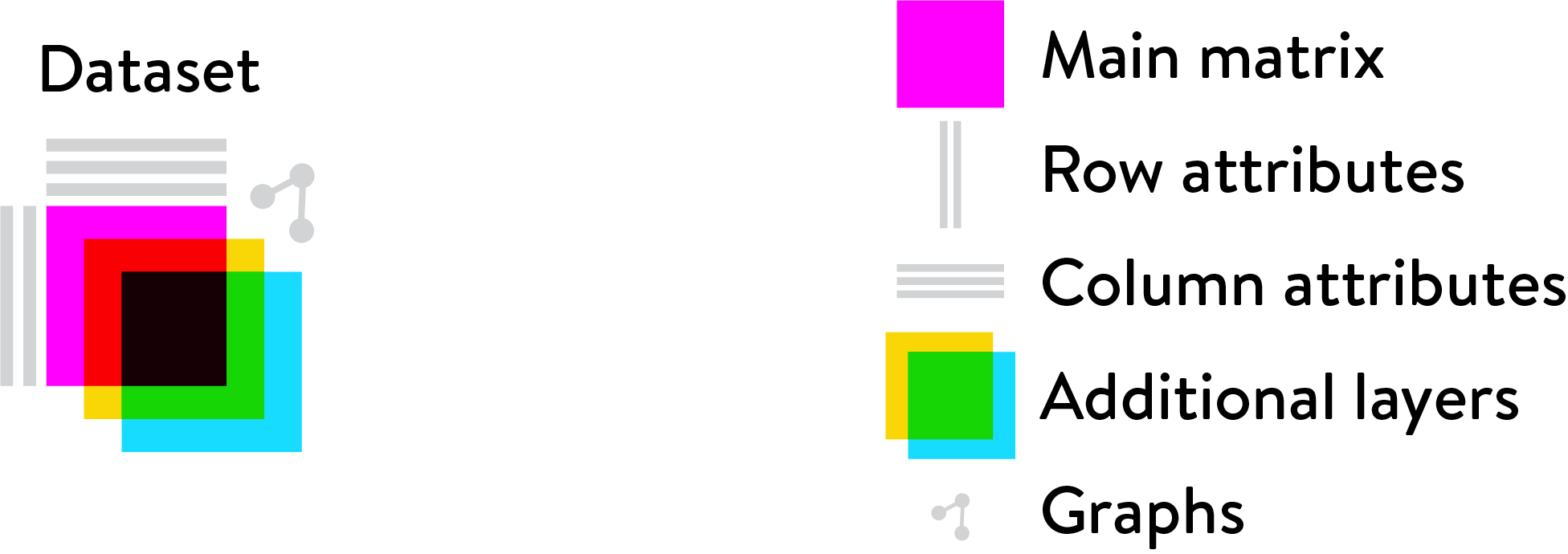
Loom files to store single-cell gene expression data: the main matrix contains the actual expression values (one column per cell, one row per gene); row and column annotations contain metadata for genes and cells, such as Name, Chromosome, Position (for genes), and Strain, Sex, Age (for cells).