SCCAF explained
Single Cell Clustering Assessment Framework (SCCAF) is a novel method for automated identification of putative cell types from single cell RNA-seq (scRNA-seq) data. By iteratively applying clustering and a machine learning approach to gene expression profiles of a given set of cells, SCCAF simultaneously identifies distinct cell groups and a weighted list of feature genes for each group. The feature genes, which are overexpressed in the particular cell group, jointly discriminate the given cell group from other cells. Each such group of cells corresponds to a putative cell type or state, characterised by the feature genes as markers.
Inputs
- AnnData object which contains the expression matrix and pre-calculated coordinates for UMAP. The AnnData object can include already clustering data, in which case the user will need to know on which AnnData slot/label is contained.
- Optional external text file with mappings between cells and clusters (when no clustering is given inside the AnnData file).
Modes of operation
- Optimisation with exit condition based on accuracy cut-off. In this case the user provides a minimum cut-off for the accuracy to be achieved and the optimisation process will exit at that point.
- Optimisation with exit condition based on under-clustered scenario. In the case the AnnData object given must include a low resolution, under clustered clustering, and its label must be know to be specified.
- Assesment only (no optimisation), where an existing clustering is assessed.
This is resource intensive.
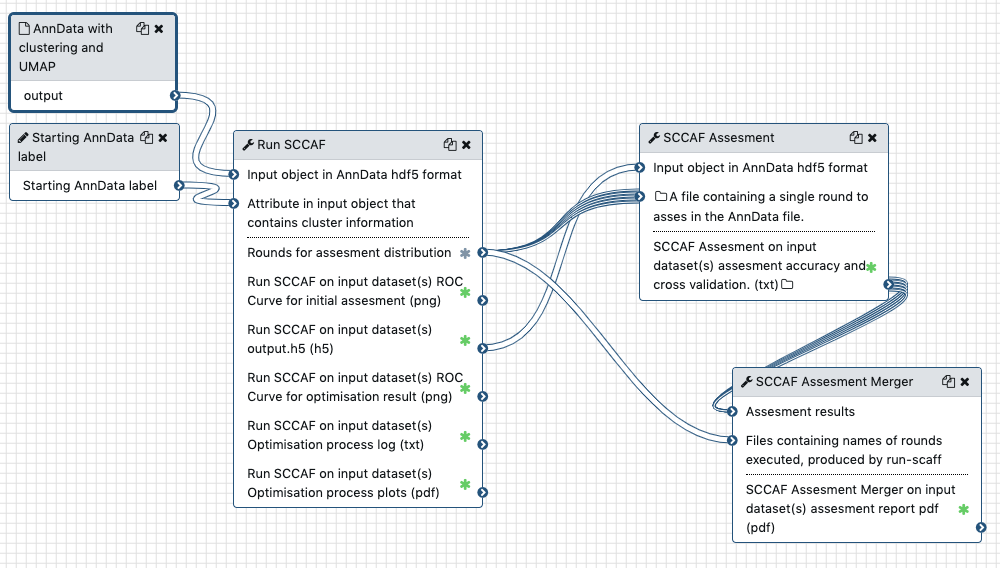
Distributed assesment
If running the optimisation you can distribute assessments of the optimisation results. For this, activate the "Produce parameter walk for assessment distribution" option, which will generate a "Rounds for assesment distribution". Then feed the AnnData output of the optimisation process and the rounds output to the SCCAF Assesment module. Then merge all assessment results with SCCAF Assesment Merger (this also receives the rounds output). The workflow would look like this: