Authors Manon Martin (manon.martin@uclouvain.be) and Marie Tremblay-Franco (marie.tremblay-franco@inra.fr; Galaxy integration)
Contact support@workflow4metabolomics.org for any questions or concerns about the Galaxy implementation of this tool.
NMR Read
Description
Nuclear Magnetic Resonance Bruker files reading (from the PEPS-NMR R package (https://github.com/ManonMartin/PEPSNMR))
Workflow position
Upstream tools
| Name | output file | format |
|---|---|---|
| NA | NA | NA |
Downstream tools
| Name | Output file | Format |
|---|---|---|
| NMR_Preprocessing | dataMatrix | Tabular |
| NMR_Preprocessing | sampleMetadata | Tabular |
| NMR_Preprocessing | NMR_Read_log | TXT |
| NMR_Preprocessing | NMR_Read_graph | |
| NMR_Alignement | dataMatrix | Tabular |
| NMR_Bucketing | dataMatrix | Tabular |
| Normalization | dataMatrix | Tabular |
| Univariate | variableMetadata | Tabular |
| Multivariate | sampleMetadata | Tabular |
| variableMetadata | Tabular |
Input files
| Parameter : num + label | Format |
|---|---|
| 1 : Choose your inputs | zip |
Choose your inputs
Zip file (recommended) of FID Bruker files: you can put a zip file containing your FID Bruker files: myinputs.zip.
Parameters
- FID Title line
- Line in the acqus file to find the FID title (name)
- subdirectories
- Organization of individual's filesTRUE: will search inside subdirectories for FIDs and will merge them to have unique FID and info matrices.
- dirs_names
- Use the (sub)directories names as FID names?
Output files
- NMR_Read_dataMatrix
- tabular outputData matrix with n rows (samples) and p columns (time) containing the raw FIDs.
- NMR_Read_sampleMetadata
- tabular outputData matrix with n rows (samples) containing the acquisition parameters for each sample.
- NMR_Read_log
- Text outputContains warnings
- NMR_Read_graph
- pdf outputline plots of FID
Creating the zip file
Must contain at least the following files for every sample: fid, acqu and acqus
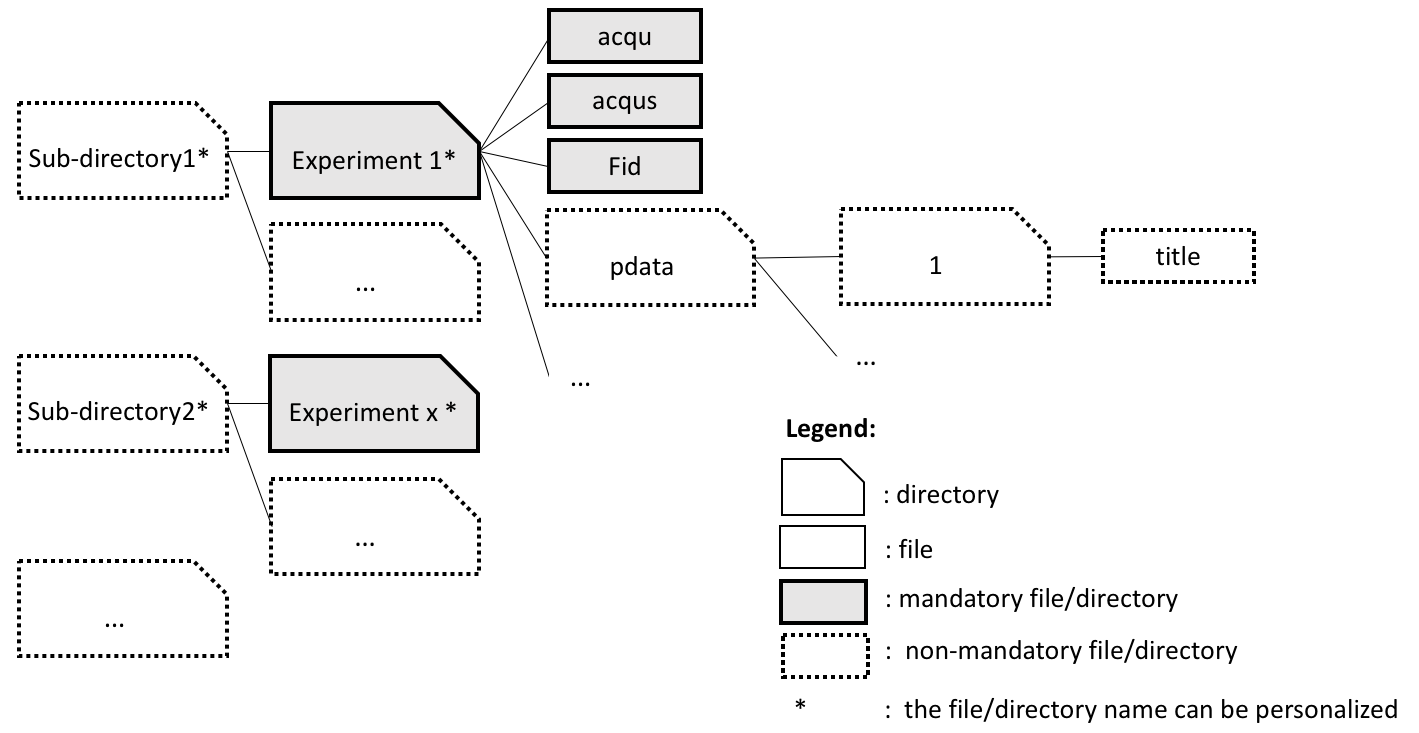
Possible structure and parameters values:
- use title file and presence of sub-directories: set the FID Title line, subdirectories = TRUE, dirs_names = FALSE
- use title file and no sub-directories: set the FID Title line, subdirectories = FALSE, dirs_names = FALSE
- don't use title file and presence of sub-directories: subdirectories = TRUE, dirs_names = TRUE
- don't use title file and no sub-directories: subdirectories = FALSE, dirs_names = TRUE
Changelog/News
Version 3.3.0
- Debugged R scripts for possibly negative baselines
Version 3.2.0
- Updated R scripts and the help section of NMR_ReadFids and NMR_preprocessing
- Added sections
- Created a variableMetadata file in output of the NMR_preprocessing module
- Homogenised the input-output file names with the other modules
- Suppressed the option ptw for the baseline correction, set to TRUE all the time now
- Shift referencing: other values than 0 are admitted
- The log file recovers the input/output parameters
- Added an exclusion zone for the computation of the Baseline Correction criterion
- Switched Internal Referencing (goes second) and Zero Order Phase Correction (goes first)
Version 3.1.0
- Implementation of NMR_ReadFids and NMR_preprocessing