Author Etienne Thevenot (W4M Core Development Team, MetaboHUB Paris, CEA)
Tool updates
See the NEWS section at the bottom of this page
Check Format
Description
Workflow position
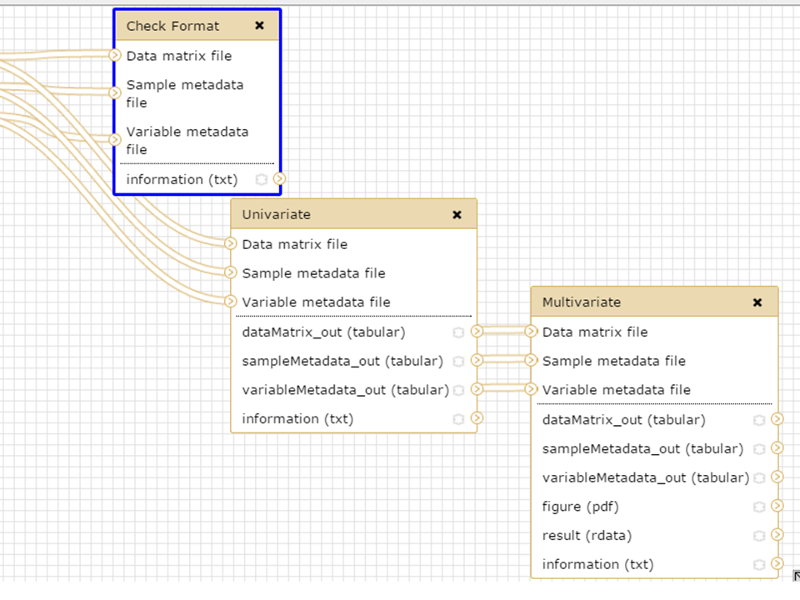
Input files
| Parameter : num + label | Format |
|---|---|
| 1 : Data matrix file | tabular |
| 2 : Sample metadata file | tabular |
| 3 : Variable metadata file | tabular |
Parameters
- Data matrix file
- variable x sample dataMatrix tabular separated file of the numeric data matrix, with . as decimal, and NA for missing values; the table must not contain metadata apart from row and column names; the row and column names must be identical to the rownames of the sample and variable metadata, respectively (see below)
- Sample metadata file
- sample x metadata sampleMetadata tabular separated file of the numeric and/or character sample metadata, with . as decimal and NA for missing values
- Variable metadata file
- variable x metadata variableMetadata tabular separated file of the numeric and/or character variable metadata, with . as decimal and NA for missing values
- Make syntactically valid sample and variable names
- if set to 'yes', sample and variable names will converted to syntactically valid names with the 'make.names' R function when required (e.g. an 'X' is added to names starting with a digit, blanks will be converted to '.', etc.)
Output files
- dataMatrix_out.tabular
- dataMatrix data file; may be identical to the input dataMatrix in case no renaming of sample/variable names nor re-ordering of samples/variables (see the 'information' file for the presence/absence of modifications)
- sampleMetadata_out.tabular
- sampleMetadata data file; may be identical to the input sampleMetadata in case no renaming of sample names nor re-ordering of samples (see the 'information' file for the presence/absence of modifications)
- variableMetadata_out.tabular
- variableMetadata data file; may be identical to the input variableMetadata in case no renaming of variable names nor re-ordering of variables (see the 'information' file for the presence/absence of modifications)
- information.txt
- Text file with all messages when error(s) in formats are detected
Working example
See the W4M00001a_sacurine-subset-statistics, W4M00001b_sacurine-complete, W4M00002_mtbls2, or W4M00003_diaplasma shared histories in the Shared Data/Published Histories menu (https://galaxy.workflow4metabolomics.org/history/list_published)
NEWS
CHANGES IN VERSION 3.0.0
NEW FEATURES
Automated re-ordering (if necessary) of sample and/or variable names from dataMatrix based on sampleMetadata and variableMetadata
New argument to make sample and variable names syntactically valid
Output of dataMatrix, sampleMetadata, and variableMetadata files, whether they have been modified or not
CHANGES IN VERSION 2.0.4
INTERNAL MODIFICATIONS
Minor internal modifications
CHANGES IN VERSION 2.0.2
INTERNAL MODIFICATIONS
Test for R code Planemo running validation Planemo installing validation Travis automated testing Toolshed export