What it does
bedtools flank will optionally create flanking intervals whose size is user-specified fraction of the original interval.
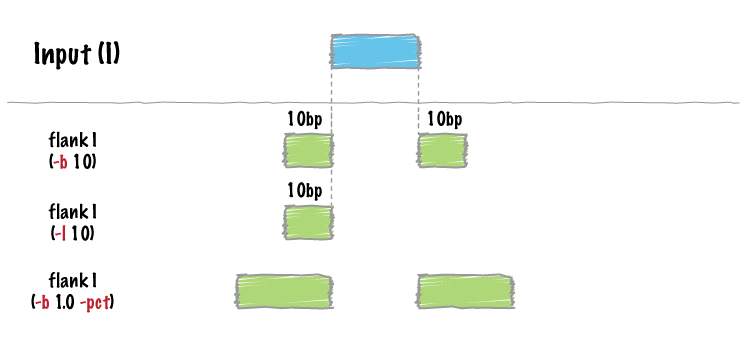
In order to prevent creating intervals that violate chromosome boundaries, bedTools flank requires a bedTool genome file defining the length of each chromosome or contig. . This should be a two column tabular file with the chromosome name in the first column and the END coordinate of the chromosome in the second column.
If you need this data for any genome that is at UCSC (http://genome.ucsc.edu), it can be extracted from the Table Browser with the "Get Data: UCSC Main" tool. Set "group" to "All Tables", "table" to "chromInfo", and "output format" to "all fields from selected table".
This tool is part of the bedtools package from the Quinlan laboratory.
Citation
If you use this tool in Galaxy, please cite:
Bjoern A. Gruening (2014), Galaxy wrapper