What it does
This program generates plots and computes table matrix of coefficient correlations and p-values at multiple scales for the correlation between the occurrences of features in one dataset and their occurrences in another using multiscale wavelet analysis technique.
The program assumes that the user has two sets of DNA sequences, S1 and S1, each of which consists of one or more sequences of equal length. Each sequence in each set is divided into the same number of multiple intervals n such that n = 2^k, where k is a positive integer and k >= 1. Thus, n could be any value of the set {2, 4, 8, 16, 32, 64, 128, ...}. k represents the number of scales.
The program has two input files obtained as follows:
For a given set of features, say motifs, the user counts the number of occurrences of each feature in each interval of each sequence in S1 and S1, and builds two tabular files representing the count results in each interval of S1 and S1. These are the input files of the program.
The program gives two output files:
- The first output file is a TABULAR format file representing the coefficient correlations and p-values for each feature at each scale.
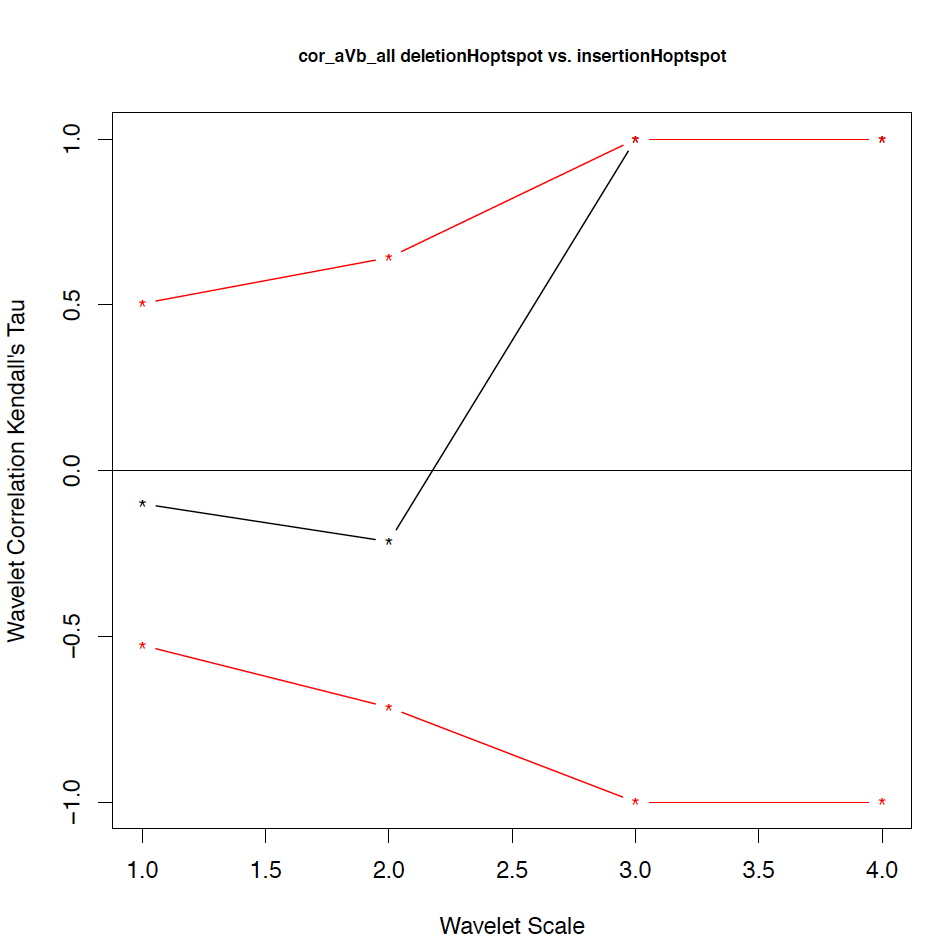
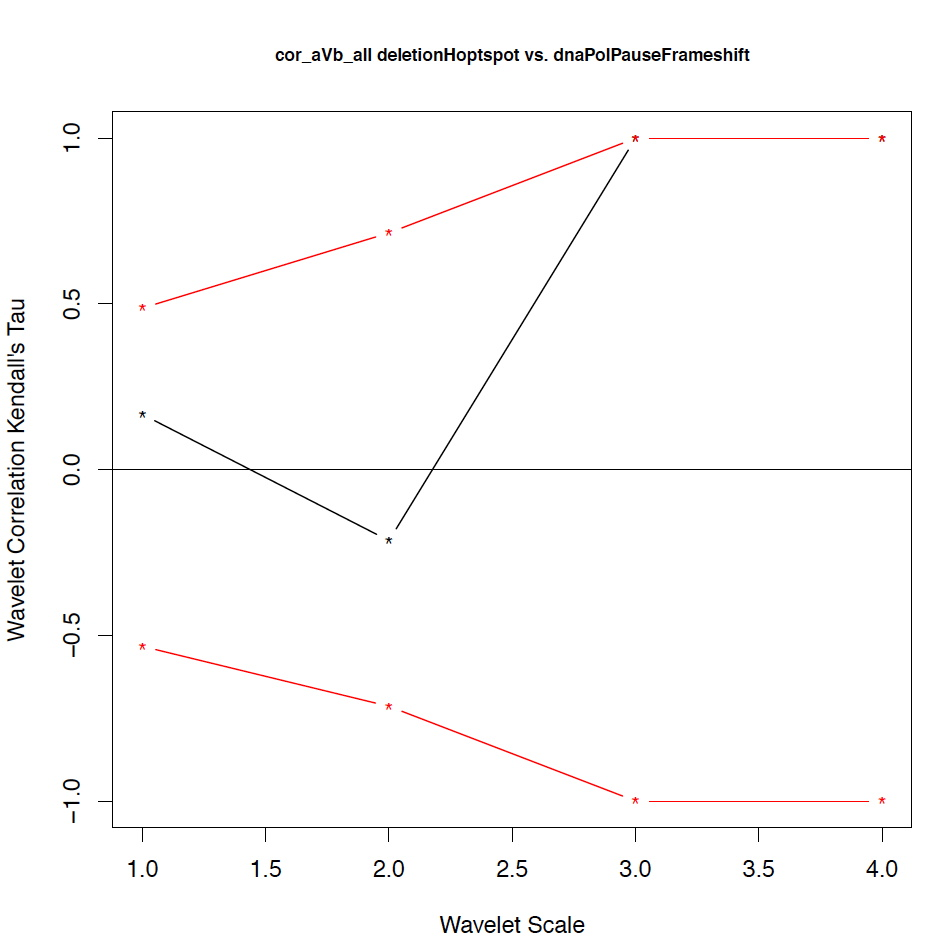
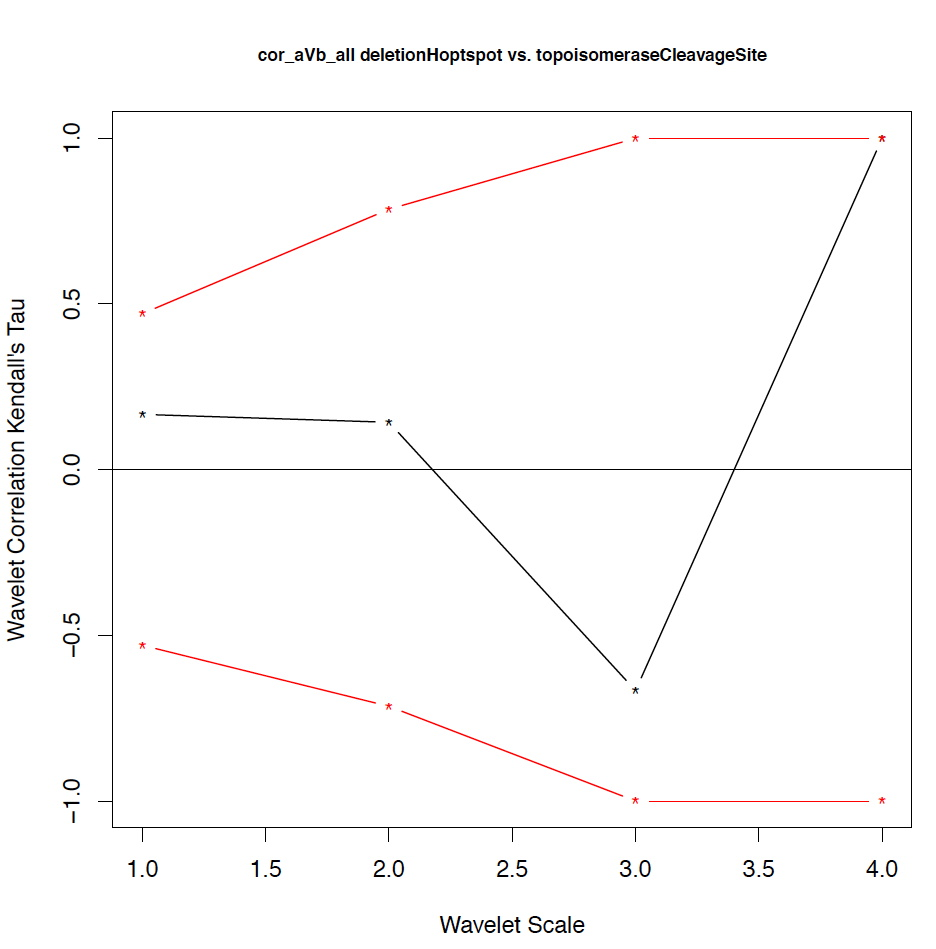
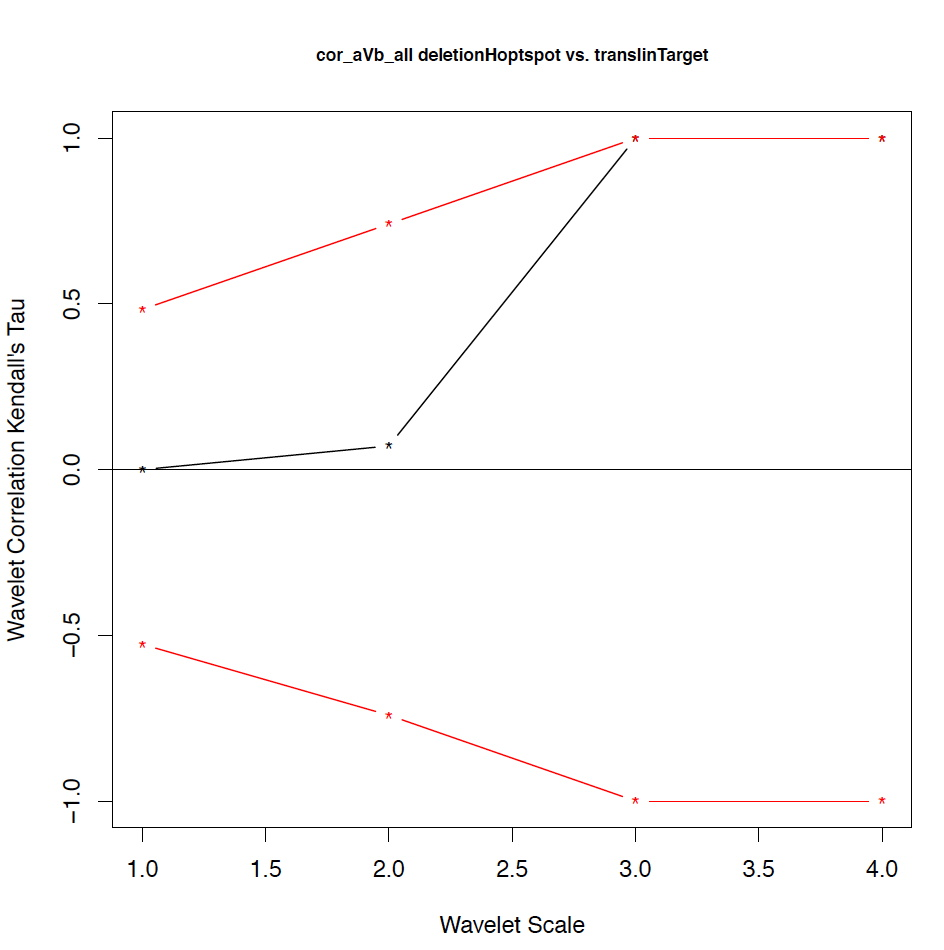
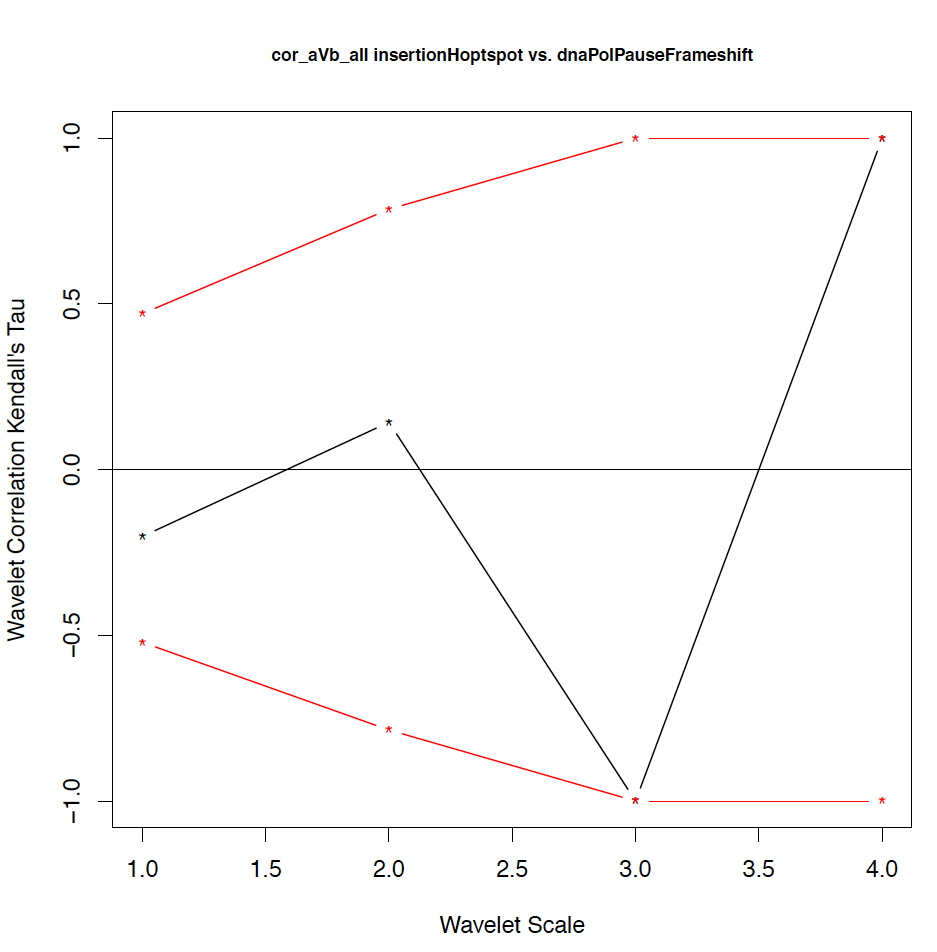
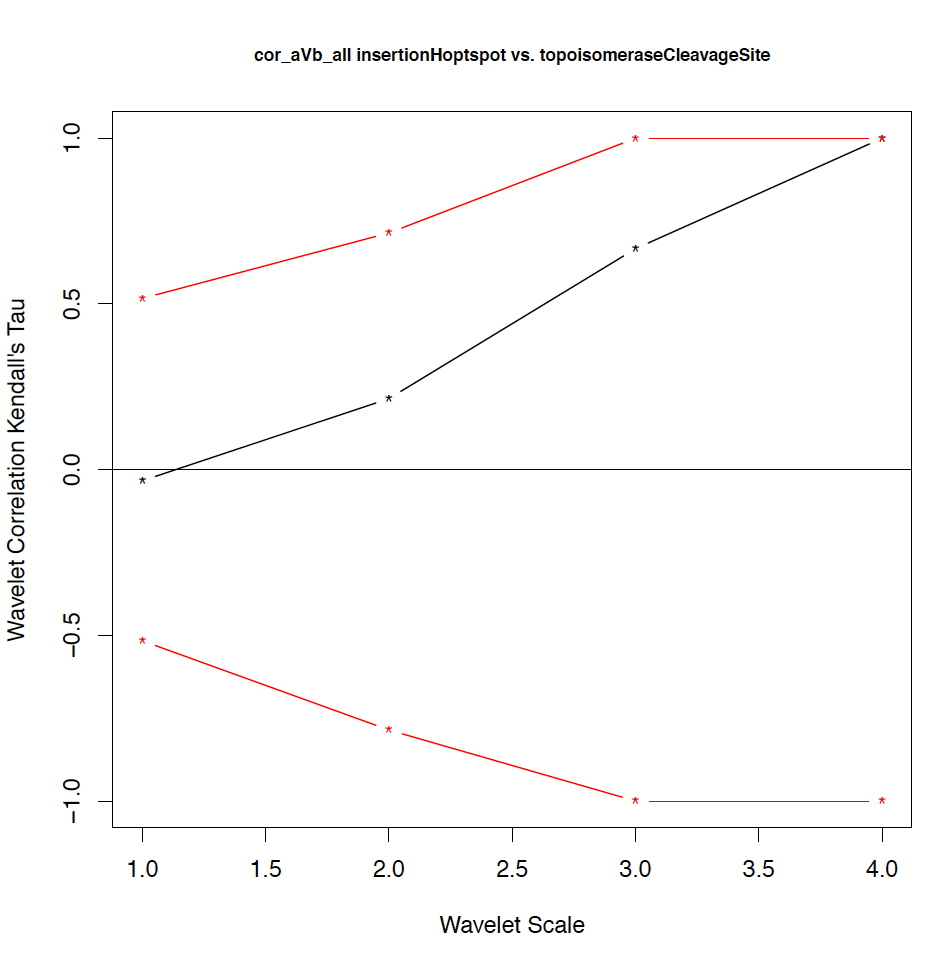
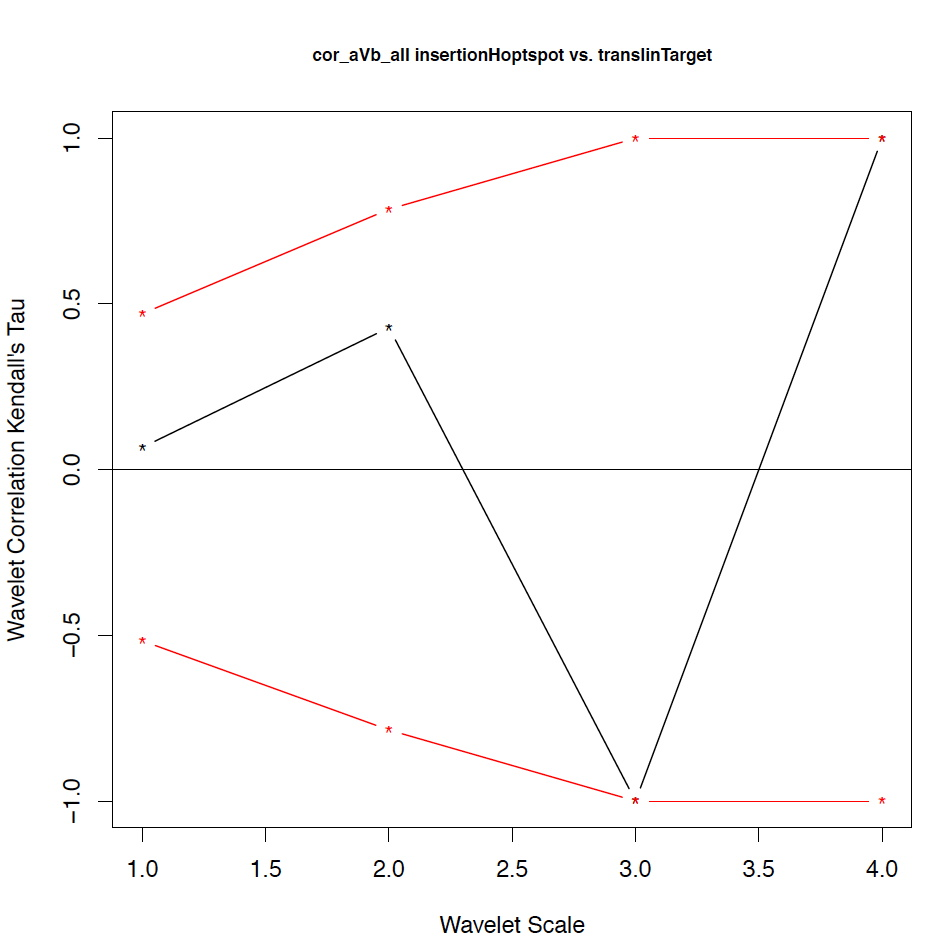
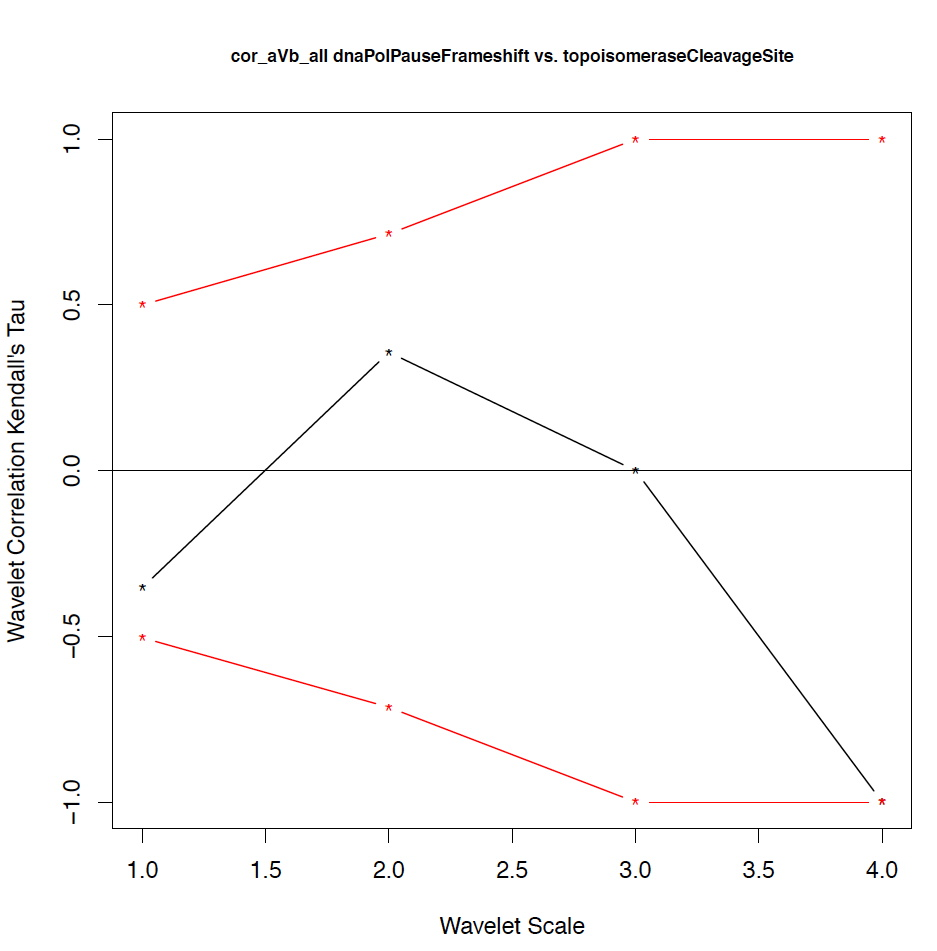
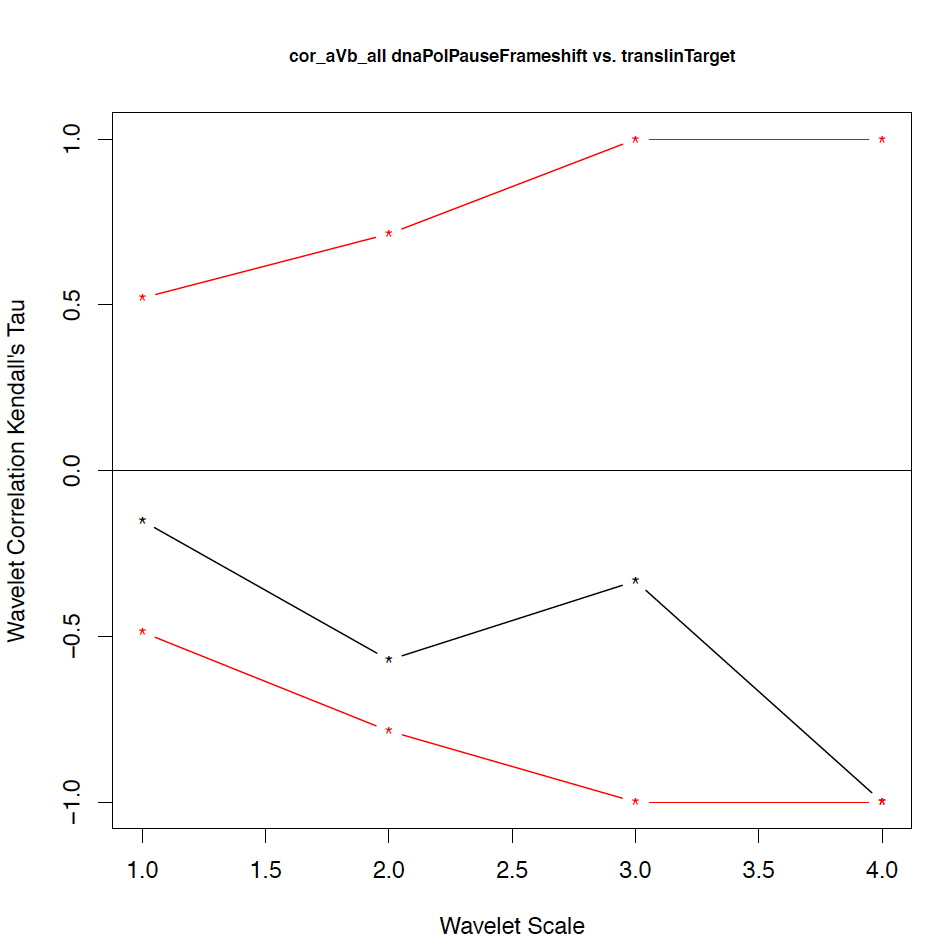
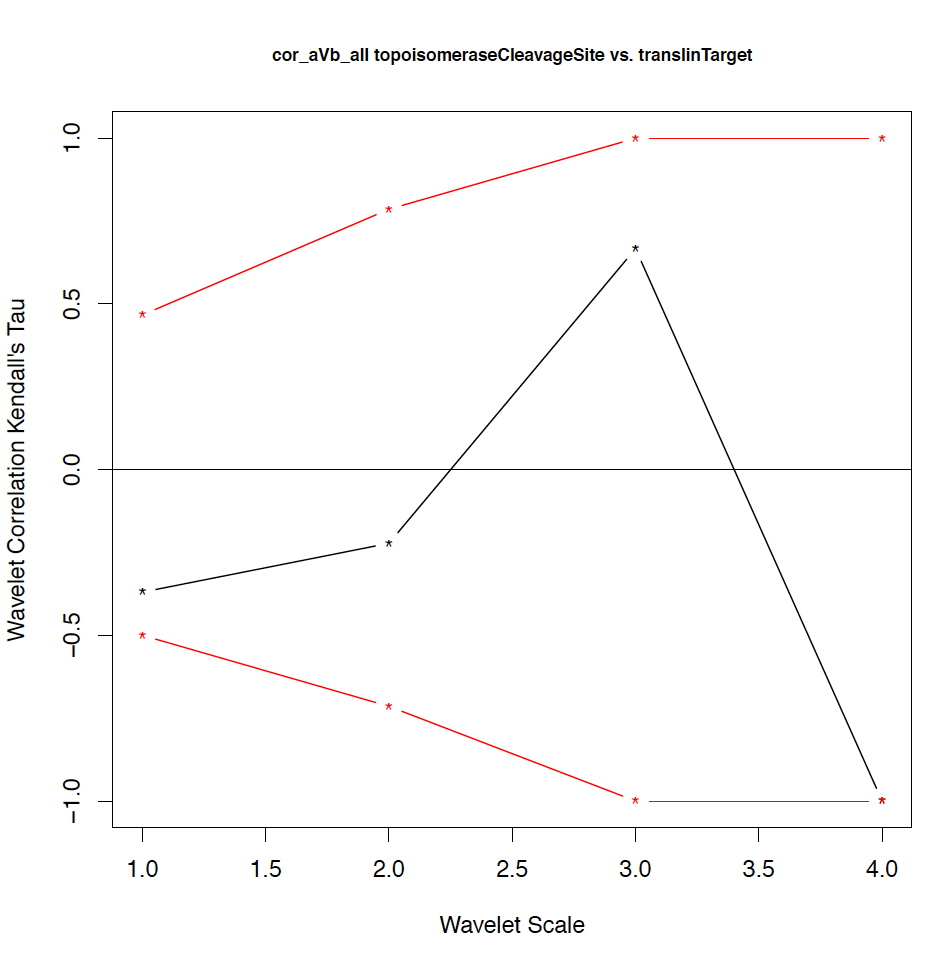
- The second output file is a PDF file consisting of as many figures as the number of features, such that each figure represents the values of the coefficient correlations for that feature at every scale.
Note
In order to obtain empirical p-values, a random perumtation test is implemented by the program, which results in the fact that the program gives slightly different results each time it is run on the same input file.
Example
Counting the occurrences of 5 features (motifs) in 16 intervals (one line per interval) of the DNA sequences in S1 gives the following tabular file:
deletionHoptspot insertionHoptspot dnaPolPauseFrameshift topoisomeraseCleavageSite translinTarget
82 162 158 79 459
111 196 154 75 459
98 178 160 79 475
113 201 170 113 436
113 173 147 95 446
107 150 155 84 436
106 166 175 96 448
113 176 135 106 514
113 170 152 87 450
95 152 167 93 467
91 171 169 118 426
84 139 160 100 459
92 154 164 104 440
100 145 154 98 472
91 161 152 71 461
117 164 139 97 463
And counting the occurrences of 5 features (motifs) in 16 intervals (one line per interval) of the DNA sequences in S2 gives the following tabular file:
deletionHoptspot insertionHoptspot dnaPolPauseFrameshift topoisomeraseCleavageSite translinTarget
269 366 330 238 1129
239 328 327 283 1188
254 351 358 297 1151
262 371 355 256 1107
254 361 352 234 1192
265 354 367 240 1182
255 359 333 235 1217
271 389 387 272 1241
240 305 341 249 1159
272 351 337 257 1169
275 351 337 233 1158
305 331 361 253 1172
277 341 343 253 1113
266 362 355 267 1162
235 326 329 241 1230
254 335 360 251 1172
We notice that the number of scales here is 4 because 16 = 2^4. Running the program on the above input files gives the following output:
The first output file:
motif1 motif2 1_cor 1_pval 2_cor 2_pval 3_cor 3_pval 4_cor 4_pval deletionHoptspot insertionHoptspot -0.1 0.346 -0.214 0.338 1 0.127 1 0.467 deletionHoptspot dnaPolPauseFrameshift 0.167 0.267 -0.214 0.334 1 0.122 1 0.511 deletionHoptspot topoisomeraseCleavageSite 0.167 0.277 0.143 0.412 -0.667 0.243 1 0.521 deletionHoptspot translinTarget 0 0.505 0.0714 0.441 1 0.124 1 0.518 insertionHoptspot dnaPolPauseFrameshift -0.202 0.238 0.143 0.379 -1 0.122 1 0.517 insertionHoptspot topoisomeraseCleavageSite -0.0336 0.457 0.214 0.29 0.667 0.252 1 0.503 insertionHoptspot translinTarget 0.0672 0.389 0.429 0.186 -1 0.119 1 0.506 dnaPolPauseFrameshift topoisomeraseCleavageSite -0.353 0.101 0.357 0.228 0 0.612 -1 0.49 dnaPolPauseFrameshift translinTarget -0.151 0.303 -0.571 0.09 -0.333 0.37 -1 1 topoisomeraseCleavageSite translinTarget -0.37 0.077 -0.222 0.297 0.667 0.234 -1 0.471
The second output file: