- Authors
- Jean-Francois Martin - PF MetaToul-AXIOM ; INRAE ; MetaboHUB (for original version of this tool and overall development of the R script)Melanie Petera - PFEM ; INRAE ; MetaboHUB (for R wrapper and R script improvement regarding "linear/lowess/loess" methods)Marion Landi - FLAME ; PFEM (for original xml interface and R wrapper)Franck Giacomoni - PFEM ; INRAE ; MetaboHUB (for original xml interface and R wrapper)Etienne Thevenot - LIST/LADIS ; CEA ; MetaboHUB (for R script and wrapper regarding "all loess pool" and "all loess sample" methods)
Please cite If you use this tool, please cite:
- when using the linear, lowess or loess methods:
- when using the all loess pool or all loess sample method:
- Cleveland et al (1997). In Statistical Models in S; Chambers JM. and Hastie TJ. Ed.; Chapman et Hall: London; pp. 309-376Etienne A. Thevenot, Aurelie Roux, Ying Xu, Eric Ezan, and Christophe Junot (2015). Analysis of the human adult urinary metabolome variations with age, body mass index and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. Journal of Proteome Research, 14:3322-3335 (http://dx.doi.org/10.1021/acs.jproteome.5b00354).
Tool updates
See the NEWS section at the bottom of this page
Batch_correction
Description
Workflow position
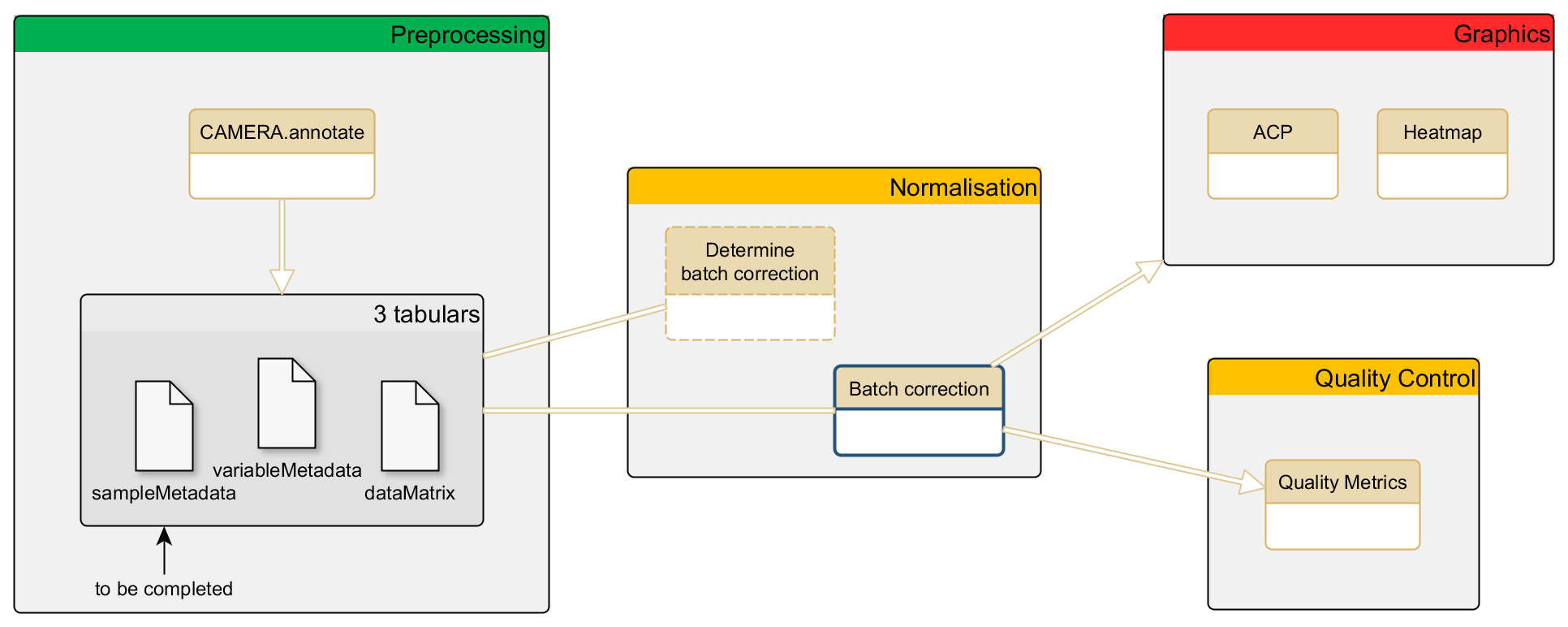
Input files
| Parameter : num + label | Format |
|---|---|
| 1 : Data Matrix file | tabular |
| 2 : Sample metadata file | tabular |
| 3 : Variable metadata file | tabular |
- Data Matrix file must contain the intensity values of variables.
- First line must contain all the samples' namesFirst column must contain all the variables' ID
- Sample metadata file must contain at least the three following columns:
- - a batch column (default to "batch") to identify the batches of analyses- an injection order column (default to "injectionOrder") composed of integers defining the injection order of samples- a sample type column (default to "sampleType") indicating if a sample is a biological one ("sample"), a QC-pool ("pool") or a blank ("blank")Default values can be changed according to your data coding using the customisation parameters in the "Sample metadata file coding parameters" section.
- Notes concerning your design:
- - the 3 mandatory columns must not contain NA- your data should contain at least 3 QC-pools in each batch for intra-batch linear adjustment and 8 for lo(w)ess adjustment (minimum of 5 for all loess methods)
Parameters
- Sample metadata file coding parameters
- Enables to give the names of columns in the sample metadata table that contain the injection order, the batches and the sample types.Also enables to specify the sample type coding used in the sampletype column.
- Type of regression model
- To choose between linear, lowess, loess, all loess pool, and all loess sample strategies- Option 1 (linear, lowess, and loess methods): before the normalisation of each variable, some quality metrics are computed (see the "Determine Batch Correction" module); depending on the result, the variable can be normalized or not, with either the linear, lowess or loess model.- Option 2 (all loess pool and all loess sample): each variable is normalized by using the 'loess' model;in the case all loess pool is chosen and the number of pool observations is below 5, the linear method is used (for all variables) and a warning is generated;if the pool intensities are not representative of the samples (which can be viewed on the figure where both trends are shown), the case all loess sample enables using the sample intensities (instead of the pool intensities) as the reference for the loess curve.In all "option 2" cases: the median intensity of the reference observations (either 'pool' or 'sample') is used as the scaling factor after the initial intensities have been divided by the loess predictions.
- Span
- Smoothing parameter, advanced option for lo(w)ess and all loess methodsIn case of a loess fit, the span parameter (between 0 and 1) controls the smoothing(the higher the smoother; higher values are prefered to avoid overfitting; Cleveland et al, 1997).
- Unconsistant values
- available for regression model linear, lowess and loessControls what is done regarding negative or infinite values that can be generated during regression estimation.Prevent it will change the normalisation term leading to an unconsistant value to prevent it:when intra batch denominator term is below 1, it is turned to the minimum >1 one obtained in the concerned batch.Consider it as a missing value will switch concerned intensities to NA;this option implies that concerned ions will not be considered in PCA display.Consider it as a null intensity will switch concerned intensities to 0.
- Factor of interest
- available for regression model linear, lowess and loessName of the factor (column header) that will be used as a categorical variable for design plots (often a biological factor ; if none, put the batch column name).This factor does not affect correction calculation.
- Level of details for plots
- available for regression model linear, lowess and loessbasic: Sum of intensities + PCA + CV boxplot (before and after correction)standard: 'basic' plots + before/after-correction plots of intensities over injection order, and design effects for each ioncomplete: 'standard' plots + QC-pool regression plots per batch with samples' intensities over injection orderThis factor is not used by the all loess methods where a unique figure is generated showing the sum of intensities along injection order, and the first 4 PCA scores.
Output files
- Batch_correction_$method_rdata.rdata ('all_loess' methods only)
- binary dataDownload, open R and use the 'load' function; objects are in the 'res' list
- Batch_correction_$method_graph.pdf
- graphical outputFor the linear and lo(w)ess methods, content depends on level of details chosen
- Batch_correction_$method_variableMetadata.tabular
- tsv outputIdentical to the variable metadata input file, with x more columns (where x is the number of batches) in case of linear, lowess and loess methods
- Batch_correction_$method_dataMatrix.tabular
- tsv output (tabulated)Same formatting as the data matrix file; contains corrected intensities
Additional information
- Refer to the corresponding "W4M HowTo" page:
- See also the reference history:
NEWS
CHANGES IN VERSION 3.0.0
NEW FEATURES
INTERNAL MODIFICATIONS
CHANGES IN VERSION 2.2.4
INTERNAL MODIFICATIONS
Fixed bug for pool selection ("all_loess" methods)
CHANGES IN VERSION 2.2.2
INTERNAL MODIFICATIONS
Fixed bug for color plot ("all_loess" methods)
CHANGES IN VERSION 2.2.0
NEW FEATURE
Specific names for the 'sampleType', 'injectionOrder', and 'batch' from sampleMetadata can be selected by the user (for compatibility with the MTBLS downloader)
CHANGES IN VERSION 2.1.2
INTERNAL MODIFICATIONS
Minor modifications in config file
CHANGES IN VERSION 2.1.0
INTERNAL MODIFICATIONS
For PCA figure display only (all_loess options): missing values are set to the minimum value before PCA computation is performed (with svd)
Additional running and installation tests added with planemo, conda, and travis
BUG FIX
Variables with NA or 0 values in all reference samples are discarded before applying the all_loess normalization
INTERNAL MODIFICATIONS
Modifications of the all_loess_wrapper file to handle the recent ropls package versions (i.e. 1.3.15 and above) which use S4 classes