Authors Eric Fontanillas created the version 1 of this pipeline. Victor Mataigne developped version 2.
Galaxy integration Julie Baffard and ABiMS TEAM, Roscoff Marine Station
Contact support.abims@sb-roscoff.fr for any questions or concerns about the Galaxy implementation of this tool.Credits : Gildas le Corguillé, Misharl Monsoor
Description
This tool takes Orthogroups found by OrthoFinder and proceeds to retrieve nucleic sequences back, then write each orthogroups in its own fasta file.
Step 1 : re-writing headers
This tool is configured to work within the AdaptSearch toolsuite, which implies a specific format of headers. Indeed, due to the use of external tools (like TransDecoder), the genes IDs might have been modified. A regular expression rewrites all the genes IDs in order to have, for instance, this format : >Pf1004_1/1_1.000_369
Step 2 : reading an re-writing orthoGroups from OrthoFinder
OrthoFinder (Emms, D.M. and Kelly, S., 2015) is a fast, accurate and comprehensive analysis tool for comparative genomics. It finds orthologues and orthogroups infers gene trees for all orthogroups and infers a rooted species tree for the species being analysed. OrthoFinder also provides comprehensive statistics for comparative genomic analyses.
Our tool focus on the orthogroups.txt file created before gene trees.The script proceeds to split each orthogroup in its own fasta file and, with the use of the output of Filter_Assemblies, to re-associate each ID with its sequence.
Inputs and parameters
- File of Orthogroups : the orthogroups.txt file from OrthoFinder.
- Output from Filter_Assemblies (either as multiple datasets or dataset collection)
- Minimal number of sequences : The orthogroups with less than the specified number won't be recorded.
- Verbose : If 'Yes', a supplementary table will be displayed in the outputs (coutings of species and sequences in orthogroups before the removal of paralogs).
- Paralogs : if 'Yes', there will be a supplementary output of orthogroups file, before the removal of paralogs.
Outputs
- Dataset collection of fasta files : each file represents an orthogroup, each gene within tthe group has the couple ID-nucleic sequence.
- If the --paralogs option is checked : another dataset collection of fasta files with all the paralogous genes.
- The tool log.
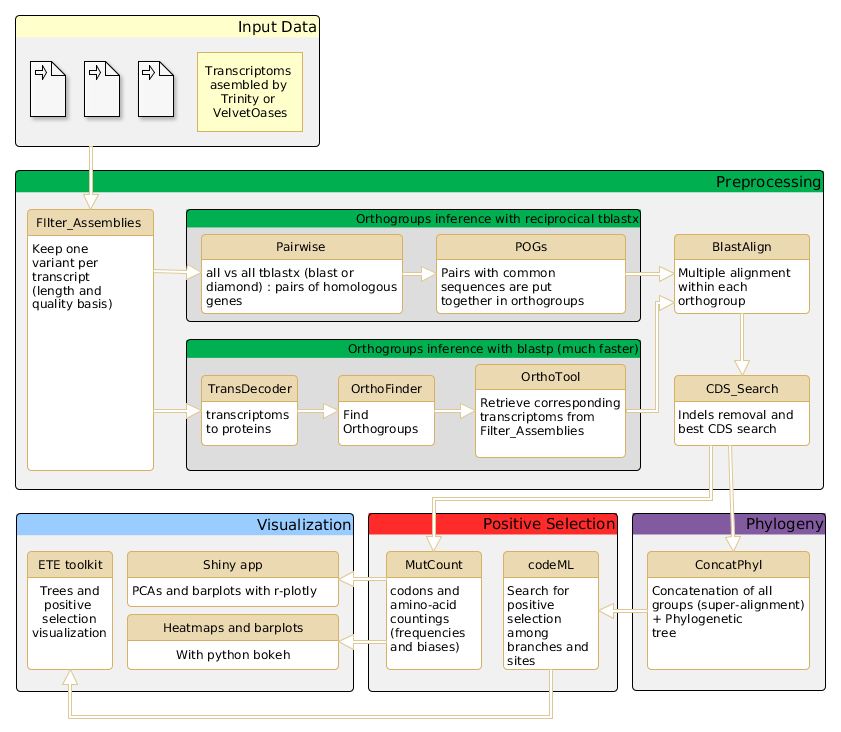
The AdaptSearch Pipeline
Changelog
Version 1.0 - 11/01/2018