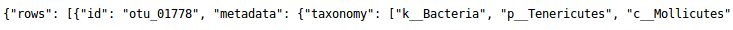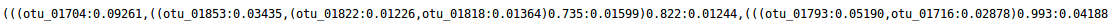What it does
Launch Rmarkdown script to import data from 3 files: biomfile, samplefile, treefile into a phyloseq object.
Inputs/Outputs
Input
OTU biom file:
The OTU biom file (format biom1). This file is the result of FROGS BIOM to std BIOM. The example of biom file:
Newick file (tree.nwk):
Newick file (format nxh) is the result of FROGS Tree:
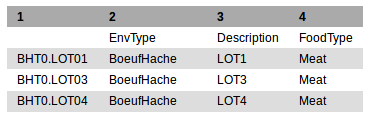
Sample file: The file contains the conditions of experiment with sample ID in the first column:
Output
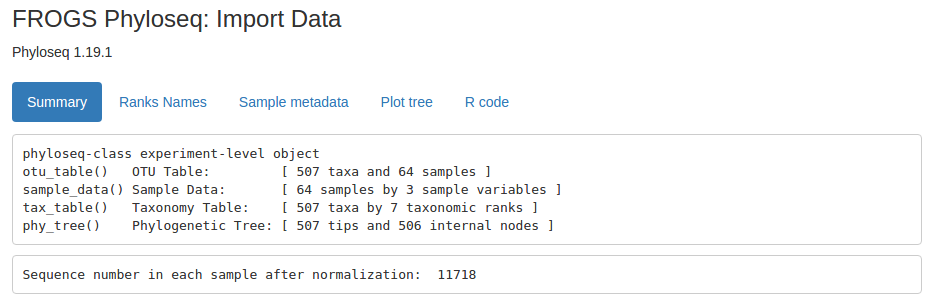
Html file (format HTML): The summary of phyloseq object.
Data file (format rdata): The informations of data in one phyloseq object.
Contact
Contacts: frogs@inra.fr
Repository: https://github.com/geraldinepascal/FROGS website: http://frogs.toulouse.inra.fr/
Please cite the FROGS article: Escudie F., et al. Bioinformatics, 2018. FROGS: Find, Rapidly, OTUs with Galaxy Solution.